Decoding the Genome of Bacteria Capable of Removing Fluorine from Persistent Perfluoro Pollutants. [View all]
The paper to which I'll briefly refer is this one: Genome-Wide Expression Analysis Unravels Fluoroalkane Metabolism in Pseudomonas sp. Strain 273 Yongchao Xie, Diana Ramirez, Gao Chen, Guang He, Yanchen Sun, Fadime Kara Murdoch, and Frank E. Löffler Environmental Science & Technology 2023 57 (42), 15925-15935.
One of the major environmental problems we face - and future generations will face for a very long time - is persistent halogenated organic molecules in the environment, the most intractable of which are the fluorinated analogues that were used to make everything from fabric protectors ("Scotch Guard"![]() to fire fighting foams, to nonstick pans to lubricants and many other products. Collectively these classes of molecules are known as "PFAS" for perfluorinated alkylated substances.
to fire fighting foams, to nonstick pans to lubricants and many other products. Collectively these classes of molecules are known as "PFAS" for perfluorinated alkylated substances.
The carbon-fluorine bond is one of the strongest in chemistry, roughly 485 kJ mol-1, which when translated to light energy puts it in the UV region for cleavage, accounting for its persistence.
I have a critic here, one of those antinuke "solar will save us" fools who fears nuclear energy more than he, she or they fears fossil fuels, even though fossil fuels people continuously without stop and nuclear energy, um, doesn't and who clearly doesn't give a rat's ass about climate change, but who (clock being right twice a day) remarked, offering up the old saw, that I am an example of one of those people when one sees a hammer as the best tool available, every problem is a nail.
As it happens I do believe that exposure to high energy radiation, available from used nuclear fuels, represents a possible way to address fluorocarbon contamination of the air, water and land, and spend a lot of time dreaming up continuous flow systems to do just that as a side product of industrial processes, but such a process would take many many generations to make significant inroads, and will be thus subject to Bateman type equilibria. It is also true that the there are some areas of contamination and contaminated matrices that cannot be exposed to high energy radiation.
Thus a living system that can get into these hard to reach places and mineralize PFAS, and despite the high energy of the bonds, there are apparently microorganisms that can metabolize fluorinated substances. That's what this paper covers.
From the introduction:
The strength of the C–F bond has been portrayed as the major hindrance to transform fluorinated organic compounds. Contrary to the belief that enzyme systems cannot cleave C–F bonds, a number of studies have reported the microbial degradation of fluorinated organic compounds. (10,11) Microbial enzyme systems can break the C–F bond through oxygenolytic, (12) hydrolytic, (13) reductive, (14) and hydration (15) mechanisms at neutral pH and at room temperature. Despite this progress, the understanding of the taxonomic diversity of microbes and their enzyme systems involved in defluorination dwarfs in comparison to the existing knowledge base about microbial dechlorination.
Pseudomonas sp. strain 273 was reported to utilize C7 to C10 1-fluoroalkanes and 1,10-difluorodecane (DFD) as the sole sources of carbon and energy for growth under oxic conditions. (16) During growth with fluoroalkanes, this bacterium releases inorganic fluoride, albeit not in stoichiometric amounts, and a fraction of organofluorine is incorporated into cellular phospholipids. (17) Growth experiments determined that the utilization of fluoroalkanes is oxygen-dependent, but the enzyme(s) responsible for defluorination have remained elusive. In this study, we integrated genomic and comparative transcriptomic investigations to identify genes that Pseudomonas sp. strain 273 utilizes during growth with DFD...
It is not clear however that the organism can eat secondary fluorine atoms, surely a major limitation, although one can imagine designing a modified protein that might accomplish this task.
The cute cartoon from the abstract:
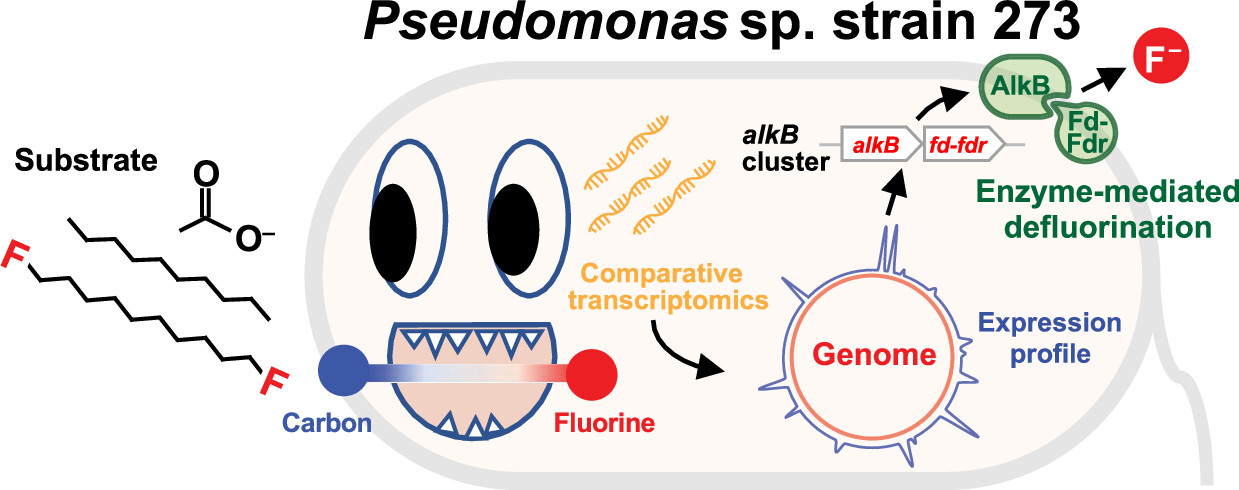
One of the more serious graphics in the paper:
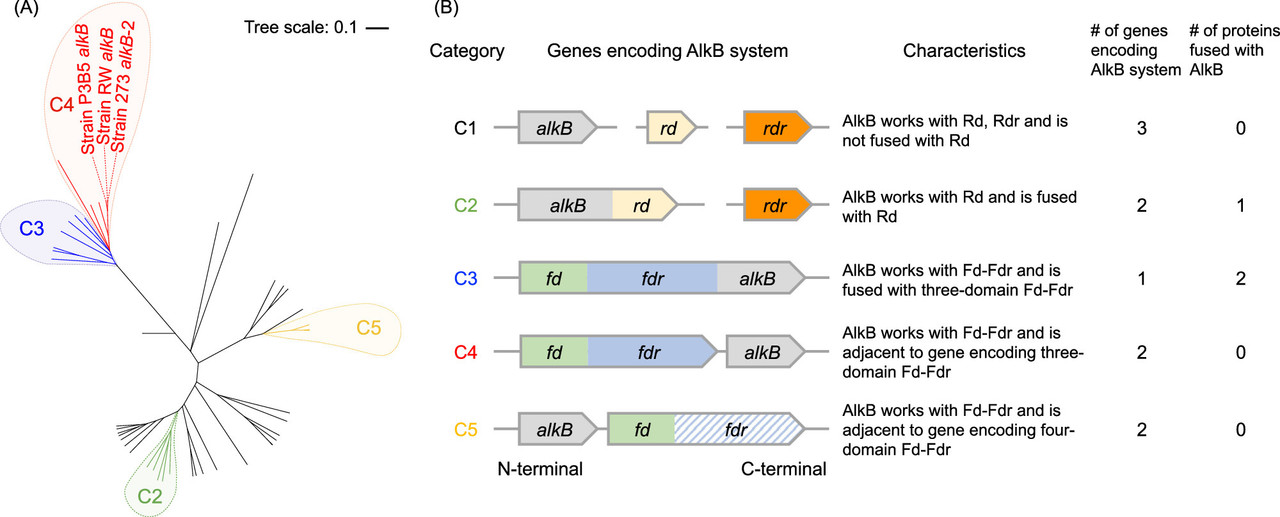
The caption:
I kind of wonder if the authors explored the defluorination of trifluoracetic acid, a penultimate product of decomposition of longer chain perfluorocarboxylic acids, and a molecule that, albeit that it occurs to a small extent naturally, is increasingly accumulating in the environment.
I trust you're enjoying the weekend.